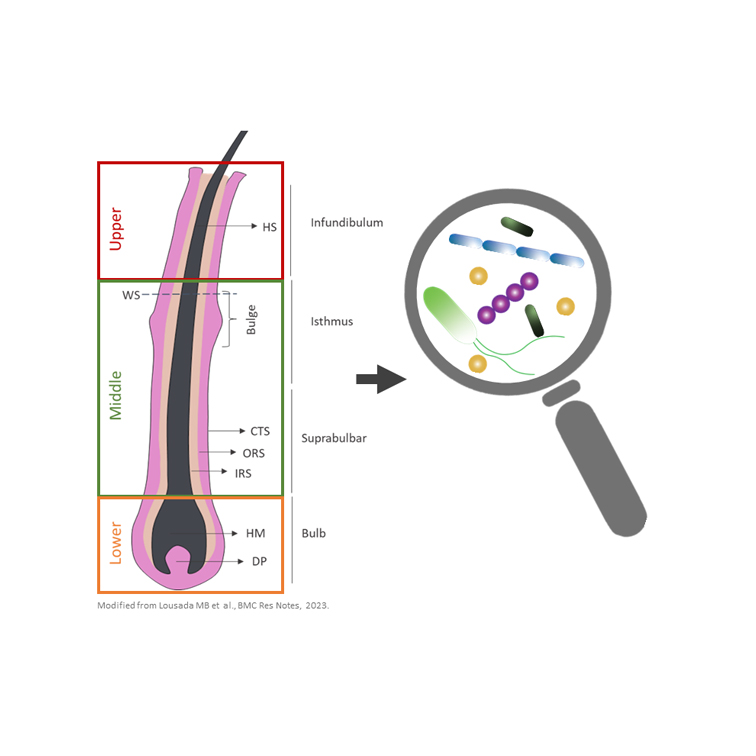
Hair follicle (HF) disorders such as acne vulgaris and hidradenitis suppurativa, also known as acne inversa, develop in response to dysregulations (imbalances) of the HF microbiota, i.e. the community of microorganisms that inhabit the human HF. Research on the HF microbiome composition is increasing and knowledge about the distinct types of microbiota present in healthy human HFs is growing. However, most of the current knowledge on the HF microbial community was generated from a skin-centric perspective with methods that do not allow to analyze distinct HF compartments, miss microbiota located in deeper HF regions or lead to contamination by extrafollicular microbial input (e.g. from the sebaceous and eccrine glands, adipocytes (fat cells), and the skin surface).
To overcome those drawbacks we developed a novel, comprehensive method where laser-capture microdissection was coupled with metagenomic techniques, which was recently published in the scientific journal BMC Research Notes. Our newly established method offers a high-resolution technique that not only allows for specific evaluation of HF tissues in their entirety, but also enables discrimination between anatomically distinct HF areas, known to be affected differently between hair disorders.
We used HFs microdissected from scalp follicular unit extractions, a technique established at Monasterium Laboratory, in combination with laser-capture microdissection technology and metagenomic 16S rRNA sequencing to generate differentiated profiles of distinct HF regions of the human scalp HF bacterial community. Interestingly we could show that – although the composition of this bacterial community is similar to the skin surface – their abundances in HFs are lower than previously reported. Yet, we confirmed that several skin populating bacteria are also present in the HF, especially Cutibacterium members, which are usually associated with sebaceous skin sites and acne vulgaris pathogenesis, were predominant. Additionally, the presence of Staphylococcus, Streptococcus, Acinetobacter and Corynebacterium was confirmed in the human HF. Besides known skin populating bacteria we also detected environmental bacteria whose presence has neither been described for the human skin nor HFs before. These microbes and their role for skin and hair function deserve further investigation. Notably, our methodological study also demonstrates for the first time that variations in the microbial profile between different HF regions (lower, middle and upper HF) exist. These differences are primarily driven by changes in abundances of Staphylococci, Streptococci, Sphingomonas and Reyranella. The findings of our study hold great translational potential because they can serve as possible starting point for the development of microbiota-based therapeutics.
We are convinced that our newly established method further allows to isolate and investigate HF compartments with an even higher resolution to specifically obtain for example mesenchymal and epithelial tissues (i.e. bulge, bulb, dermal papilla, connective tissue sheath). This information could be used to generate a detailed microbial map of healthy and/or diseased human HFs to understand the dysbiotic events occurring in several hair disorders. Thus, the findings from our pilot study strongly suggest our method as an invaluable tool for the development of microbiota-based therapeutics to treat dysbiosis-associated hair disorders.
Read more about it here: Laser capture microdissection as a method for investigating the human hair follicle microbiome reveals region-specific differences in the bacteriome profile
The study was mainly performed by our Junior Staff Scientist Marta Lousada, in collaboration with the Zoological Institute of the University of Kiel.